Temporal clustering on a single channel¶
Run a non-parametric cluster stats on sensor EEG070 on the contrast faces vs. scrambled.
import os.path as op
import sys
import numpy as np
from scipy import stats
import matplotlib.pyplot as plt
import mne
from mne.stats import permutation_cluster_1samp_test
sys.path.append(op.join('..', '..', 'processing'))
from library.config import (meg_dir, l_freq, N_JOBS, set_matplotlib_defaults,
exclude_subjects, annot_kwargs, random_state) # noqa: E402
Read all the data
contrasts = list()
for subject_id in range(1, 20):
if subject_id in exclude_subjects:
continue
subject = "sub%03d" % subject_id
print("processing subject: %s" % subject)
data_path = op.join(meg_dir, subject)
contrast = mne.read_evokeds(op.join(data_path, '%s_highpass-%sHz-ave.fif'
% (subject, l_freq)),
condition='contrast')
contrast.apply_baseline((-0.2, 0.0)).crop(None, 0.8)
contrast.pick_types(meg=False, eeg=True)
contrasts.append(contrast)
contrast = mne.combine_evoked(contrasts, 'equal')
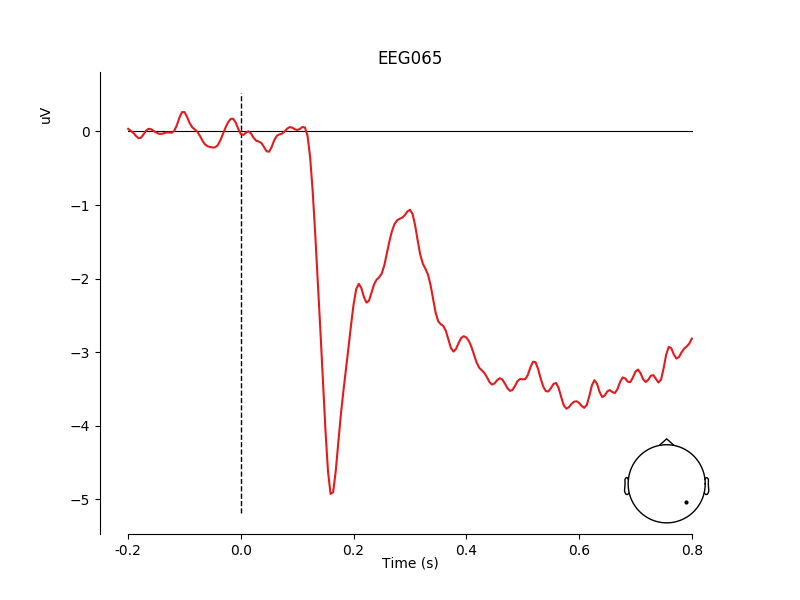
channel = 'EEG065'
idx = contrast.ch_names.index(channel)
mne.viz.plot_compare_evokeds(contrast, [idx], show_sensors=4,
truncate_xaxis=False)
Out:
processing subject: sub002
Reading /tsi/doctorants/data_gramfort/dgw_faces_reproduce/MEG/sub002/sub002_highpass-NoneHz-ave.fif ...
Read a total of 1 projection items:
Average EEG reference (1 x 70) active
Found the data of interest:
t = -200.00 ... 2900.00 ms (contrast)
0 CTF compensation matrices available
nave = 155 - aspect type = 100
Projections have already been applied. Setting proj attribute to True.
No baseline correction applied
Applying baseline correction (mode: mean)
processing subject: sub003
Reading /tsi/doctorants/data_gramfort/dgw_faces_reproduce/MEG/sub003/sub003_highpass-NoneHz-ave.fif ...
Read a total of 1 projection items:
Average EEG reference (1 x 70) active
Found the data of interest:
t = -200.00 ... 2900.00 ms (contrast)
0 CTF compensation matrices available
nave = 146 - aspect type = 100
Projections have already been applied. Setting proj attribute to True.
No baseline correction applied
Applying baseline correction (mode: mean)
processing subject: sub004
Reading /tsi/doctorants/data_gramfort/dgw_faces_reproduce/MEG/sub004/sub004_highpass-NoneHz-ave.fif ...
Read a total of 1 projection items:
Average EEG reference (1 x 70) active
Found the data of interest:
t = -200.00 ... 2900.00 ms (contrast)
0 CTF compensation matrices available
nave = 139 - aspect type = 100
Projections have already been applied. Setting proj attribute to True.
No baseline correction applied
Applying baseline correction (mode: mean)
processing subject: sub006
Reading /tsi/doctorants/data_gramfort/dgw_faces_reproduce/MEG/sub006/sub006_highpass-NoneHz-ave.fif ...
Read a total of 1 projection items:
Average EEG reference (1 x 70) active
Found the data of interest:
t = -200.00 ... 2900.00 ms (contrast)
0 CTF compensation matrices available
nave = 108 - aspect type = 100
Projections have already been applied. Setting proj attribute to True.
No baseline correction applied
Applying baseline correction (mode: mean)
processing subject: sub007
Reading /tsi/doctorants/data_gramfort/dgw_faces_reproduce/MEG/sub007/sub007_highpass-NoneHz-ave.fif ...
Read a total of 1 projection items:
Average EEG reference (1 x 70) active
Found the data of interest:
t = -200.00 ... 2900.00 ms (contrast)
0 CTF compensation matrices available
nave = 193 - aspect type = 100
Projections have already been applied. Setting proj attribute to True.
No baseline correction applied
Applying baseline correction (mode: mean)
processing subject: sub008
Reading /tsi/doctorants/data_gramfort/dgw_faces_reproduce/MEG/sub008/sub008_highpass-NoneHz-ave.fif ...
Read a total of 1 projection items:
Average EEG reference (1 x 70) active
Found the data of interest:
t = -200.00 ... 2900.00 ms (contrast)
0 CTF compensation matrices available
nave = 127 - aspect type = 100
Projections have already been applied. Setting proj attribute to True.
No baseline correction applied
Applying baseline correction (mode: mean)
processing subject: sub009
Reading /tsi/doctorants/data_gramfort/dgw_faces_reproduce/MEG/sub009/sub009_highpass-NoneHz-ave.fif ...
Read a total of 1 projection items:
Average EEG reference (1 x 70) active
Found the data of interest:
t = -200.00 ... 2900.00 ms (contrast)
0 CTF compensation matrices available
nave = 89 - aspect type = 100
Projections have already been applied. Setting proj attribute to True.
No baseline correction applied
Applying baseline correction (mode: mean)
processing subject: sub010
Reading /tsi/doctorants/data_gramfort/dgw_faces_reproduce/MEG/sub010/sub010_highpass-NoneHz-ave.fif ...
Read a total of 1 projection items:
Average EEG reference (1 x 70) active
Found the data of interest:
t = -200.00 ... 2900.00 ms (contrast)
0 CTF compensation matrices available
nave = 115 - aspect type = 100
Projections have already been applied. Setting proj attribute to True.
No baseline correction applied
Applying baseline correction (mode: mean)
processing subject: sub011
Reading /tsi/doctorants/data_gramfort/dgw_faces_reproduce/MEG/sub011/sub011_highpass-NoneHz-ave.fif ...
Read a total of 1 projection items:
Average EEG reference (1 x 70) active
Found the data of interest:
t = -200.00 ... 2900.00 ms (contrast)
0 CTF compensation matrices available
nave = 113 - aspect type = 100
Projections have already been applied. Setting proj attribute to True.
No baseline correction applied
Applying baseline correction (mode: mean)
processing subject: sub012
Reading /tsi/doctorants/data_gramfort/dgw_faces_reproduce/MEG/sub012/sub012_highpass-NoneHz-ave.fif ...
Read a total of 1 projection items:
Average EEG reference (1 x 70) active
Found the data of interest:
t = -200.00 ... 2900.00 ms (contrast)
0 CTF compensation matrices available
nave = 116 - aspect type = 100
Projections have already been applied. Setting proj attribute to True.
No baseline correction applied
Applying baseline correction (mode: mean)
processing subject: sub013
Reading /tsi/doctorants/data_gramfort/dgw_faces_reproduce/MEG/sub013/sub013_highpass-NoneHz-ave.fif ...
Read a total of 1 projection items:
Average EEG reference (1 x 70) active
Found the data of interest:
t = -200.00 ... 2900.00 ms (contrast)
0 CTF compensation matrices available
nave = 120 - aspect type = 100
Projections have already been applied. Setting proj attribute to True.
No baseline correction applied
Applying baseline correction (mode: mean)
processing subject: sub014
Reading /tsi/doctorants/data_gramfort/dgw_faces_reproduce/MEG/sub014/sub014_highpass-NoneHz-ave.fif ...
Read a total of 1 projection items:
Average EEG reference (1 x 70) active
Found the data of interest:
t = -200.00 ... 2900.00 ms (contrast)
0 CTF compensation matrices available
nave = 116 - aspect type = 100
Projections have already been applied. Setting proj attribute to True.
No baseline correction applied
Applying baseline correction (mode: mean)
processing subject: sub015
Reading /tsi/doctorants/data_gramfort/dgw_faces_reproduce/MEG/sub015/sub015_highpass-NoneHz-ave.fif ...
Read a total of 1 projection items:
Average EEG reference (1 x 70) active
Found the data of interest:
t = -200.00 ... 2900.00 ms (contrast)
0 CTF compensation matrices available
nave = 60 - aspect type = 100
Projections have already been applied. Setting proj attribute to True.
No baseline correction applied
Applying baseline correction (mode: mean)
processing subject: sub017
Reading /tsi/doctorants/data_gramfort/dgw_faces_reproduce/MEG/sub017/sub017_highpass-NoneHz-ave.fif ...
Read a total of 1 projection items:
Average EEG reference (1 x 70) active
Found the data of interest:
t = -200.00 ... 2900.00 ms (contrast)
0 CTF compensation matrices available
nave = 89 - aspect type = 100
Projections have already been applied. Setting proj attribute to True.
No baseline correction applied
Applying baseline correction (mode: mean)
processing subject: sub018
Reading /tsi/doctorants/data_gramfort/dgw_faces_reproduce/MEG/sub018/sub018_highpass-NoneHz-ave.fif ...
Read a total of 1 projection items:
Average EEG reference (1 x 70) active
Found the data of interest:
t = -200.00 ... 2900.00 ms (contrast)
0 CTF compensation matrices available
nave = 135 - aspect type = 100
Projections have already been applied. Setting proj attribute to True.
No baseline correction applied
Applying baseline correction (mode: mean)
processing subject: sub019
Reading /tsi/doctorants/data_gramfort/dgw_faces_reproduce/MEG/sub019/sub019_highpass-NoneHz-ave.fif ...
Read a total of 1 projection items:
Average EEG reference (1 x 70) active
Found the data of interest:
t = -200.00 ... 2900.00 ms (contrast)
0 CTF compensation matrices available
nave = 138 - aspect type = 100
Projections have already been applied. Setting proj attribute to True.
No baseline correction applied
Applying baseline correction (mode: mean)
Assemble the data and run the cluster stats on channel data
data = np.array([c.data[idx] for c in contrasts])
n_permutations = 1000 # number of permutations to run
# set initial threshold
p_initial = 0.001
# set family-wise p-value
p_thresh = 0.01
connectivity = None
tail = 0. # for two sided test
# set cluster threshold
n_samples = len(data)
threshold = -stats.t.ppf(p_initial / (1 + (tail == 0)), n_samples - 1)
if np.sign(tail) < 0:
threshold = -threshold
cluster_stats = permutation_cluster_1samp_test(
data, threshold=threshold, n_jobs=N_JOBS, verbose=True, tail=tail,
step_down_p=0.05, connectivity=connectivity,
n_permutations=n_permutations, seed=random_state)
T_obs, clusters, cluster_p_values, _ = cluster_stats
Out:
stat_fun(H1): min=-8.458284 max=2.145938
Running initial clustering
Found 2 clusters
Permuting 999 times...
[ ] 0.10010 |
[. ] 3.20320 /
[.. ] 6.40641 -
[... ] 9.60961 \
[..... ] 12.81281 |
[...... ] 16.01602 /
[....... ] 19.21922 -
[........ ] 22.42242 \
[.......... ] 25.62563 |
[........... ] 28.82883 /
[............ ] 32.03203 -
[.............. ] 35.23524 \
[............... ] 38.43844 |
[................ ] 41.64164 /
[................. ] 44.84484 -
[................... ] 48.04805 \
[.................... ] 51.25125 |
[..................... ] 54.45445 /
[....................... ] 57.65766 -
[........................ ] 60.86086 \
[......................... ] 64.06406 |
[.......................... ] 67.26727 /
[............................ ] 70.47047 -
[............................. ] 73.67367 \
[.............................. ] 76.87688 |
[................................ ] 80.08008 /
[................................. ] 83.28328 -
[.................................. ] 86.48649 \
[................................... ] 89.68969 |
[..................................... ] 92.89289 /
[...................................... ] 96.09610 -
[....................................... ] 99.29930 \ Computing cluster p-values
Step-down-in-jumps iteration #1 found 2 clusters to exclude from subsequent iterations
Permuting 999 times...
[ ] 0.10010 |
[. ] 3.20320 /
[.. ] 6.40641 -
[... ] 9.60961 \
[..... ] 12.81281 |
[...... ] 16.01602 /
[....... ] 19.21922 -
[........ ] 22.42242 \
[.......... ] 25.62563 |
[........... ] 28.82883 /
[............ ] 32.03203 -
[.............. ] 35.23524 \
[............... ] 38.43844 |
[................ ] 41.64164 /
[................. ] 44.84484 -
[................... ] 48.04805 \
[.................... ] 51.25125 |
[..................... ] 54.45445 /
[....................... ] 57.65766 -
[........................ ] 60.86086 \
[......................... ] 64.06406 |
[.......................... ] 67.26727 /
[............................ ] 70.47047 -
[............................. ] 73.67367 \
[.............................. ] 76.87688 |
[................................ ] 80.08008 /
[................................. ] 83.28328 -
[.................................. ] 86.48649 \
[................................... ] 89.68969 |
[..................................... ] 92.89289 /
[...................................... ] 96.09610 -
[....................................... ] 99.29930 \ Computing cluster p-values
Step-down-in-jumps iteration #2 found 0 additional clusters to exclude from subsequent iterations
Done.
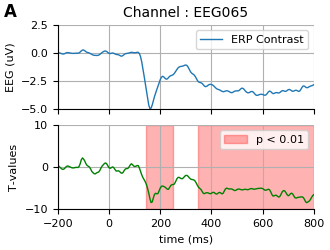
Visualize results
set_matplotlib_defaults()
times = 1e3 * contrast.times
fig, axes = plt.subplots(2, sharex=True, figsize=(3.3, 2.5))
ax = axes[0]
ax.plot(times, 1e6 * data.mean(axis=0), label="ERP Contrast")
ax.set(title='Channel : ' + channel, ylabel="EEG (uV)", ylim=[-5, 2.5])
ax.legend()
ax.annotate('A', (-0.16, 1.15), **annot_kwargs)
ax = axes[1]
for i_c, c in enumerate(clusters):
c = c[0]
if cluster_p_values[i_c] < p_thresh:
h1 = ax.axvspan(times[c.start], times[c.stop - 1],
color='r', alpha=0.3)
hf = ax.plot(times, T_obs, 'g')
ax.legend((h1,), (u'p < %s' % p_thresh,), loc='upper right', ncol=1)
ax.set(xlabel="time (ms)", ylabel="T-values",
ylim=[-10., 10.], xlim=contrast.times[[0, -1]] * 1000)
fig.tight_layout(pad=0.5)
fig.savefig(op.join('..', 'figures', 'sensorstat_highpass-%sHz.pdf'
% (l_freq,)), bbox_to_inches='tight')
plt.show()
Total running time of the script: ( 0 minutes 1.940 seconds)