Spatio-temporal source-space statistics (MEG)¶
Clustering in source space.
from functools import partial
import os.path as op
import sys
import numpy as np
from scipy import stats
from mayavi import mlab
import mne
from mne import spatial_src_connectivity
from mne.stats import (spatio_temporal_cluster_1samp_test,
summarize_clusters_stc, ttest_1samp_no_p)
sys.path.append(op.join('..', '..', 'processing'))
from library.config import (meg_dir, subjects_dir, fsaverage_vertices,
exclude_subjects, N_JOBS, l_freq, random_state) # noqa: E402
faces = list()
scrambled = list()
for subject_id in range(1, 20):
if subject_id in exclude_subjects:
continue
subject = "sub%03d" % subject_id
print("processing subject: %s" % subject)
data_path = op.join(meg_dir, subject)
stc = mne.read_source_estimate(
op.join(data_path, 'mne_dSPM_inverse_morph_highpass-%sHz-faces_eq'
% (l_freq,)))
faces.append(stc.magnitude().crop(None, 0.8).data.T)
stc = mne.read_source_estimate(
op.join(data_path, 'mne_dSPM_inverse_morph_highpass-%sHz-scrambled_eq'
% (l_freq,)))
scrambled.append(stc.magnitude().crop(None, 0.8).data.T)
tstep = stc.tstep
Out:
processing subject: sub002
processing subject: sub003
processing subject: sub004
processing subject: sub006
processing subject: sub007
processing subject: sub008
processing subject: sub009
processing subject: sub010
processing subject: sub011
processing subject: sub012
processing subject: sub013
processing subject: sub014
processing subject: sub015
processing subject: sub017
processing subject: sub018
processing subject: sub019
Set up our contrast and initial p-value threshold
X = np.array(faces, float) - np.array(scrambled, float)
fsaverage_src = mne.read_source_spaces(
op.join(subjects_dir, 'fsaverage', 'bem', 'fsaverage-5-src.fif'))
connectivity = spatial_src_connectivity(fsaverage_src)
# something like 0.01 is a more typical value here (or use TFCE!), but
# for speed here we'll use 0.001 (fewer clusters to handle)
p_threshold = 0.001
t_threshold = -stats.distributions.t.ppf(p_threshold / 2., len(X) - 1)
Out:
Reading a source space...
Computing patch statistics...
Patch information added...
Distance information added...
[done]
Reading a source space...
Computing patch statistics...
Patch information added...
Distance information added...
[done]
2 source spaces read
-- number of connected vertices : 20484
Here we could do an exact test with n_permutations=2**(len(X)-1),
i.e. 32768 permutations, but this would take a long time. For speed and
simplicity we’ll do 1024.
stat_fun = partial(ttest_1samp_no_p, sigma=1e-3)
T_obs, clusters, cluster_p_values, H0 = clu = \
spatio_temporal_cluster_1samp_test(
X, connectivity=connectivity, n_jobs=N_JOBS, threshold=t_threshold,
stat_fun=stat_fun, buffer_size=None, seed=random_state,
step_down_p=0.05, verbose=True)
good_cluster_inds = np.where(cluster_p_values < 0.05)[0]
for ind in good_cluster_inds:
print('Found cluster with p=%g' % (cluster_p_values[ind],))
Out:
stat_fun(H1): min=-5.241046 max=11.401539
Running initial clustering
Found 900 clusters
Permuting 1023 times...
[ ] 0.09775 |
[. ] 3.12805 /
[.. ] 6.25611 -
[... ] 9.38416 \
[..... ] 12.51222 |
[...... ] 15.64027 /
[....... ] 18.76833 -
[........ ] 21.89638 \
[.......... ] 25.02444 |
[........... ] 28.15249 /
[............ ] 31.28055 -
[............. ] 34.40860 \
[............... ] 37.53666 |
[................ ] 40.66471 /
[................. ] 43.79277 -
[.................. ] 46.92082 \
[.................... ] 50.04888 |
[..................... ] 53.17693 /
[...................... ] 56.30499 -
[....................... ] 59.43304 \
[......................... ] 62.56109 |
[.......................... ] 65.68915 /
[........................... ] 68.81720 -
[............................ ] 71.94526 \
[.............................. ] 75.07331 |
[............................... ] 78.20137 /
[................................ ] 81.32942 -
[................................. ] 84.45748 \
[................................... ] 87.58553 |
[.................................... ] 90.71359 /
[..................................... ] 93.84164 -
[...................................... ] 96.96970 \ Computing cluster p-values
Step-down-in-jumps iteration #1 found 7 clusters to exclude from subsequent iterations
Permuting 1023 times...
[ ] 0.09775 |
[. ] 3.12805 /
[.. ] 6.25611 -
[... ] 9.38416 \
[..... ] 12.51222 |
[...... ] 15.64027 /
[....... ] 18.76833 -
[........ ] 21.89638 \
[.......... ] 25.02444 |
[........... ] 28.15249 /
[............ ] 31.28055 -
[............. ] 34.40860 \
[............... ] 37.53666 |
[................ ] 40.66471 /
[................. ] 43.79277 -
[.................. ] 46.92082 \
[.................... ] 50.04888 |
[..................... ] 53.17693 /
[...................... ] 56.30499 -
[....................... ] 59.43304 \
[......................... ] 62.56109 |
[.......................... ] 65.68915 /
[........................... ] 68.81720 -
[............................ ] 71.94526 \
[.............................. ] 75.07331 |
[............................... ] 78.20137 /
[................................ ] 81.32942 -
[................................. ] 84.45748 \
[................................... ] 87.58553 |
[.................................... ] 90.71359 /
[..................................... ] 93.84164 -
[...................................... ] 96.96970 \ Computing cluster p-values
Step-down-in-jumps iteration #2 found 0 additional clusters to exclude from subsequent iterations
Done.
Found cluster with p=0.015625
Found cluster with p=0.0273438
Found cluster with p=0.0117188
Found cluster with p=0.0429688
Found cluster with p=0.0214844
Found cluster with p=0.00585938
Found cluster with p=0.00976562
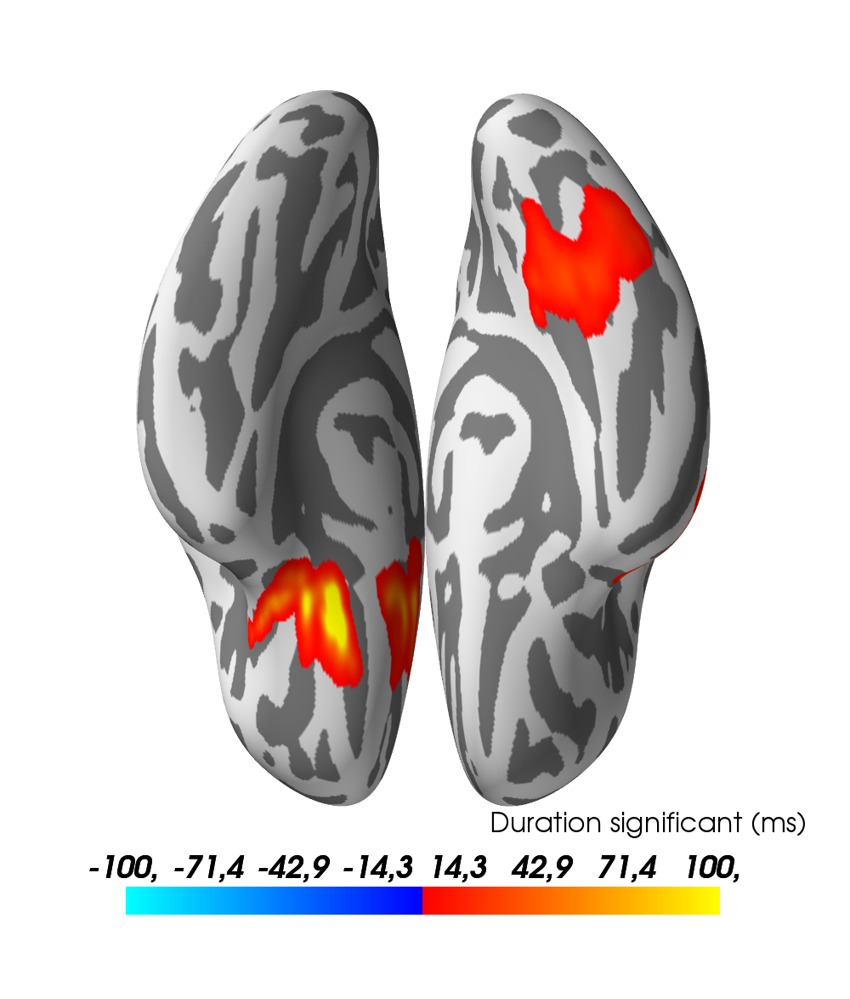
Visualize the results:
stc_all_cluster_vis = summarize_clusters_stc(
clu, tstep=tstep, vertices=fsaverage_vertices, subject='fsaverage')
pos_lims = [0, 0.1, 100 if l_freq is None else 30]
brain = stc_all_cluster_vis.plot(
hemi='both', subjects_dir=subjects_dir,
time_label='Duration significant (ms)', views='ven',
clim=dict(pos_lims=pos_lims, kind='value'), size=(1000, 1000),
background='white', foreground='black')
mlab.view(90, 180, roll=180, focalpoint=(0., 15., 0.), distance=500)
brain.save_image(op.join('..', 'figures', 'source_stats_highpass-%sHz.png'
% (l_freq,)))
Out:
Updating smoothing matrix, be patient..
Smoothing matrix creation, step 1
Smoothing matrix creation, step 2
Smoothing matrix creation, step 3
Smoothing matrix creation, step 4
Smoothing matrix creation, step 5
Smoothing matrix creation, step 6
Smoothing matrix creation, step 7
Smoothing matrix creation, step 8
Smoothing matrix creation, step 9
Smoothing matrix creation, step 10
colormap: fmin=-1.00e+02 fmid=0.00e+00 fmax=1.00e+02 transparent=0
Updating smoothing matrix, be patient..
Smoothing matrix creation, step 1
Smoothing matrix creation, step 2
Smoothing matrix creation, step 3
Smoothing matrix creation, step 4
Smoothing matrix creation, step 5
Smoothing matrix creation, step 6
Smoothing matrix creation, step 7
Smoothing matrix creation, step 8
Smoothing matrix creation, step 9
Smoothing matrix creation, step 10
colormap: fmin=-1.00e+02 fmid=0.00e+00 fmax=1.00e+02 transparent=0
setting roll
Total running time of the script: ( 40 minutes 13.008 seconds)