Analysis for subject 16¶
Run the analysis.
import os.path as op
import sys
import numpy as np
import matplotlib.pyplot as plt
import mne
sys.path.append(op.join('..', '..', 'processing'))
from library.config import (study_path, meg_dir, ylim, l_freq,
set_matplotlib_defaults) # noqa: E402
set_matplotlib_defaults()
Configuration
subjects_dir = op.join(study_path, 'subjects')
subject = "sub016"
subject_dir = op.join(meg_dir, subject)
Continuous data
raw_fname = op.join(study_path, 'ds117', subject, 'MEG', 'run_01_raw.fif')
raw_filt_fname = op.join(subject_dir,
'run_01_filt_sss_highpass-%sHz_raw.fif' % l_freq)
raw = mne.io.read_raw_fif(raw_fname)
raw_filt = mne.io.read_raw_fif(raw_filt_fname)
Out:
Opening raw data file /tsi/doctorants/data_gramfort/dgw_faces_reproduce/ds117/sub016/MEG/run_01_raw.fif...
Read a total of 8 projection items:
mag_ssp_upright.fif : PCA-mags-v1 (1 x 306) idle
mag_ssp_upright.fif : PCA-mags-v2 (1 x 306) idle
mag_ssp_upright.fif : PCA-mags-v3 (1 x 306) idle
mag_ssp_upright.fif : PCA-mags-v4 (1 x 306) idle
mag_ssp_upright.fif : PCA-mags-v5 (1 x 306) idle
grad_ssp_upright.fif : PCA-grad-v1 (1 x 306) idle
grad_ssp_upright.fif : PCA-grad-v2 (1 x 306) idle
grad_ssp_upright.fif : PCA-grad-v3 (1 x 306) idle
Range : 214500 ... 760099 = 195.000 ... 690.999 secs
Ready.
Current compensation grade : 0
Opening raw data file /tsi/doctorants/data_gramfort/dgw_faces_reproduce/MEG/sub016/run_01_filt_sss_highpass-NoneHz_raw.fif...
Range : 214500 ... 760099 = 195.000 ... 690.999 secs
Ready.
Current compensation grade : 0
Filtering 04. Filter using MNE-python.
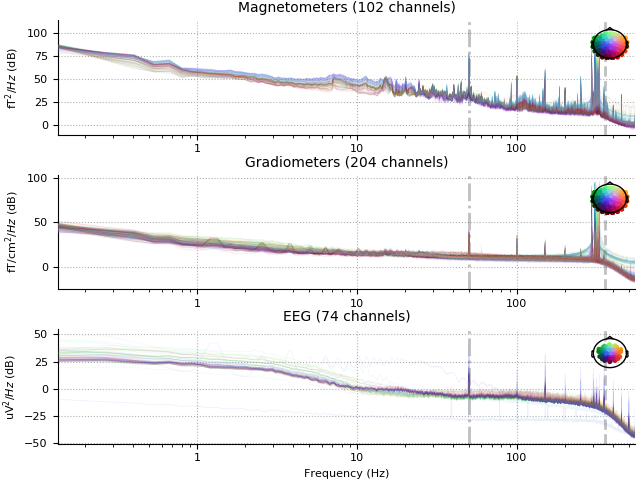
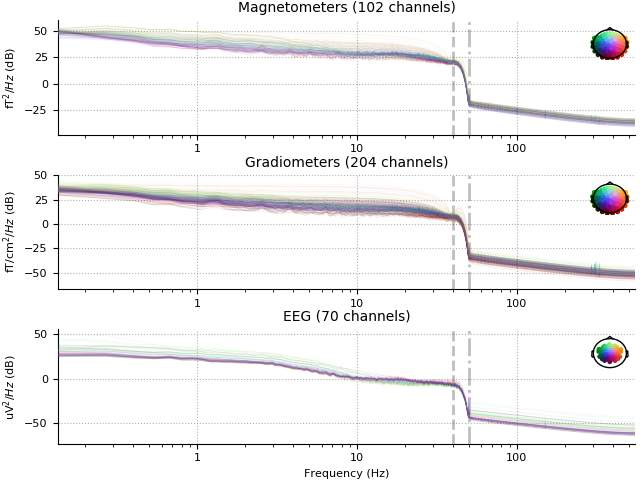
raw.plot_psd(n_fft=8192, average=False, xscale='log', show=False)
raw_filt.plot_psd(n_fft=8192, average=False, xscale='log')
Out:
Effective window size : 7.447 (s)
Effective window size : 7.447 (s)
Effective window size : 7.447 (s)
Effective window size : 7.447 (s)
Effective window size : 7.447 (s)
Effective window size : 7.447 (s)
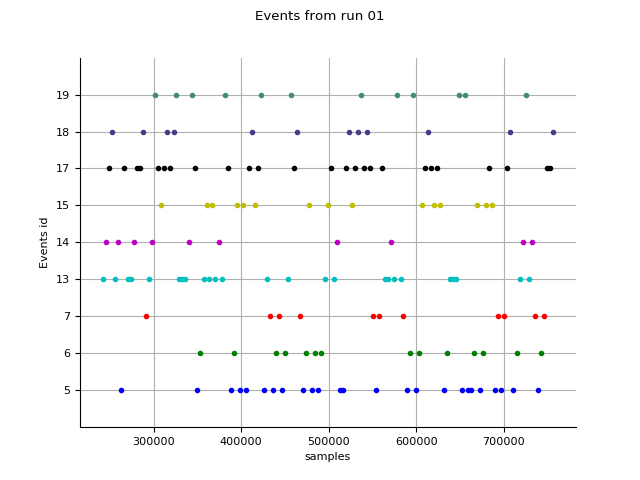
Events 02. Extract events from the stimulus channel. Epochs 06. Construct epochs.
eve_fname = op.join(subject_dir, 'run_01-eve.fif')
epo_fname = op.join(subject_dir,
'%s_highpass-%sHz-epo.fif' % (subject, l_freq))
events = mne.read_events(eve_fname)
fig = mne.viz.plot_events(events, show=False)
fig.suptitle('Events from run 01')
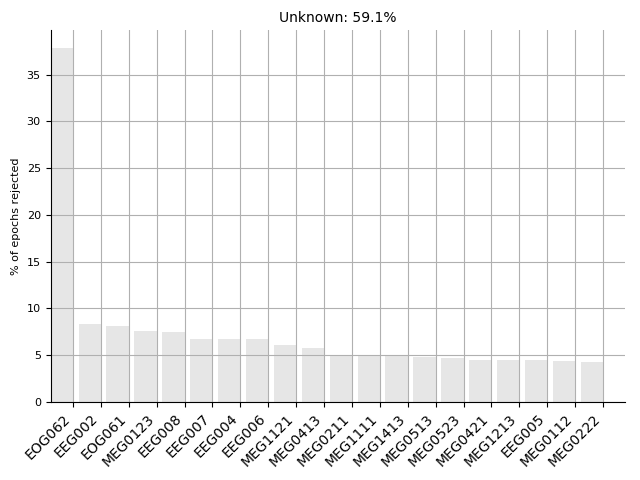
epochs = mne.read_epochs(epo_fname)
epochs.plot_drop_log()
Out:
Reading /tsi/doctorants/data_gramfort/dgw_faces_reproduce/MEG/sub016/sub016_highpass-NoneHz-epo.fif ...
Read a total of 1 projection items:
Average EEG reference (1 x 70) active
Found the data of interest:
t = -200.00 ... 2900.00 ms
0 CTF compensation matrices available
363 matching events found
Created an SSP operator (subspace dimension = 1)
363 matching events found
Created an SSP operator (subspace dimension = 1)
1 projection items activated
Evoked responses 07. Evoked data
ave_fname = op.join(subject_dir,
'%s_highpass-%sHz-ave.fif' % (subject, l_freq))
evoked = mne.read_evokeds(ave_fname)
famous_evo, scrambled_evo, unfamiliar_evo, contrast_evo, faces_evo = evoked[:5]
Out:
Reading /tsi/doctorants/data_gramfort/dgw_faces_reproduce/MEG/sub016/sub016_highpass-NoneHz-ave.fif ...
Read a total of 1 projection items:
Average EEG reference (1 x 70) active
Found the data of interest:
t = -200.00 ... 2900.00 ms (famous)
0 CTF compensation matrices available
nave = 124 - aspect type = 100
Projections have already been applied. Setting proj attribute to True.
No baseline correction applied
Read a total of 1 projection items:
Average EEG reference (1 x 70) active
Found the data of interest:
t = -200.00 ... 2900.00 ms (scrambled)
0 CTF compensation matrices available
nave = 110 - aspect type = 100
Projections have already been applied. Setting proj attribute to True.
No baseline correction applied
Read a total of 1 projection items:
Average EEG reference (1 x 70) active
Found the data of interest:
t = -200.00 ... 2900.00 ms (unfamiliar)
0 CTF compensation matrices available
nave = 129 - aspect type = 100
Projections have already been applied. Setting proj attribute to True.
No baseline correction applied
Read a total of 1 projection items:
Average EEG reference (1 x 70) active
Found the data of interest:
t = -200.00 ... 2900.00 ms (contrast)
0 CTF compensation matrices available
nave = 77 - aspect type = 100
Projections have already been applied. Setting proj attribute to True.
No baseline correction applied
Read a total of 1 projection items:
Average EEG reference (1 x 70) active
Found the data of interest:
t = -200.00 ... 2900.00 ms (faces)
0 CTF compensation matrices available
nave = 253 - aspect type = 100
Projections have already been applied. Setting proj attribute to True.
No baseline correction applied
Read a total of 1 projection items:
Average EEG reference (1 x 70) active
Found the data of interest:
t = -200.00 ... 2900.00 ms (faces_eq)
0 CTF compensation matrices available
nave = 110 - aspect type = 100
Projections have already been applied. Setting proj attribute to True.
No baseline correction applied
Read a total of 1 projection items:
Average EEG reference (1 x 70) active
Found the data of interest:
t = -200.00 ... 2900.00 ms (scrambled_eq)
0 CTF compensation matrices available
nave = 110 - aspect type = 100
Projections have already been applied. Setting proj attribute to True.
No baseline correction applied
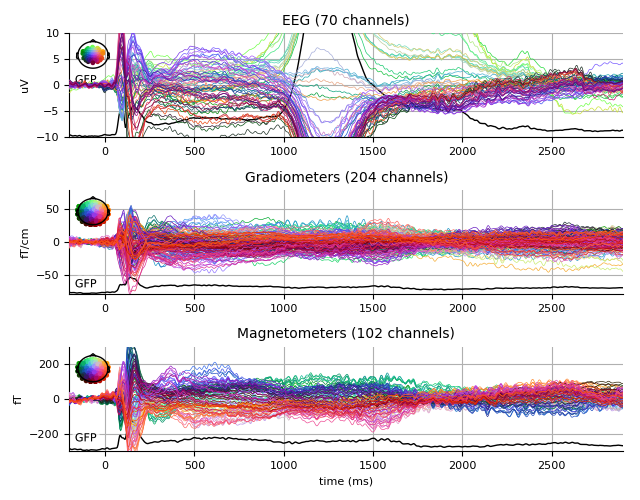
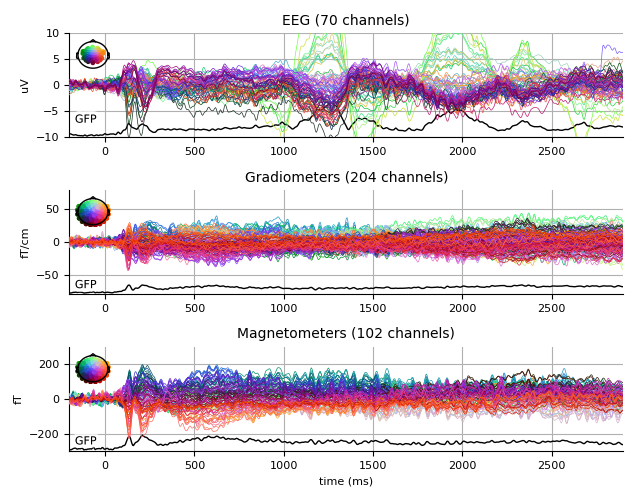
Faces
faces_evo.plot(spatial_colors=True, gfp=True, ylim=ylim,
window_title='Faces %s' % subject)
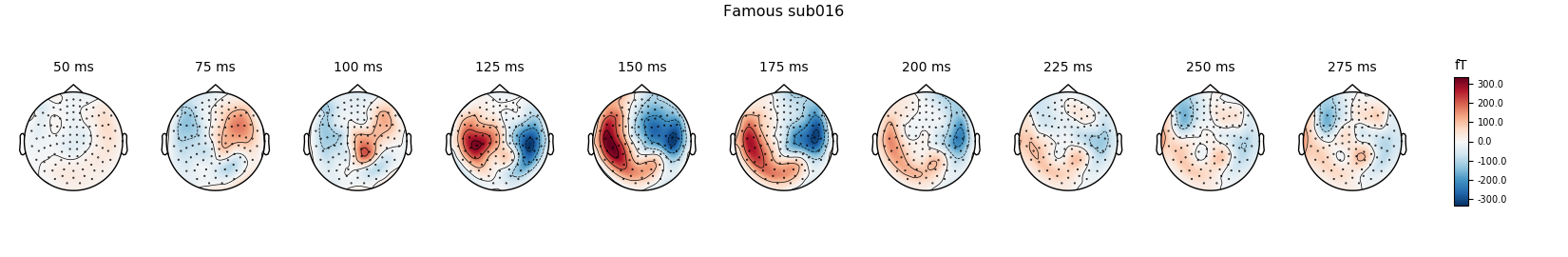
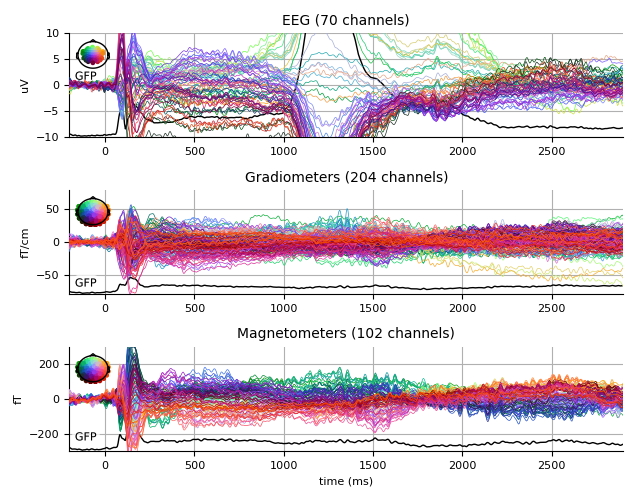
Famous
famous_evo.plot(spatial_colors=True, gfp=True, ylim=ylim,
window_title='Famous %s' % subject)
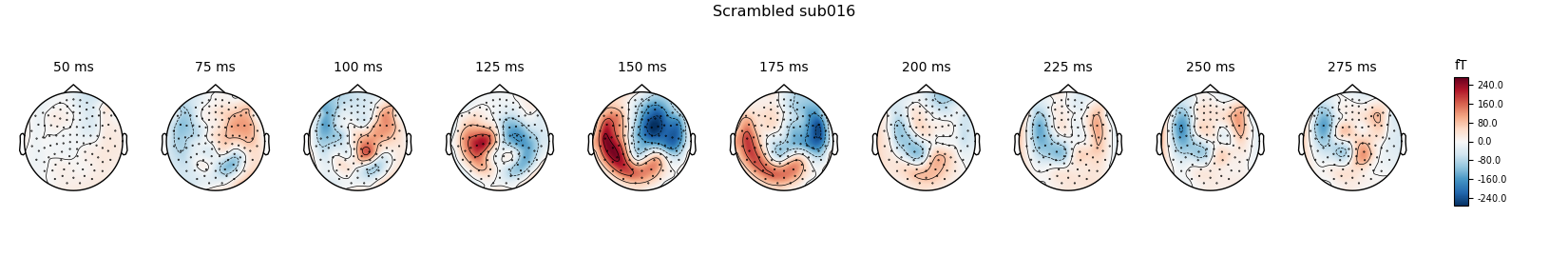
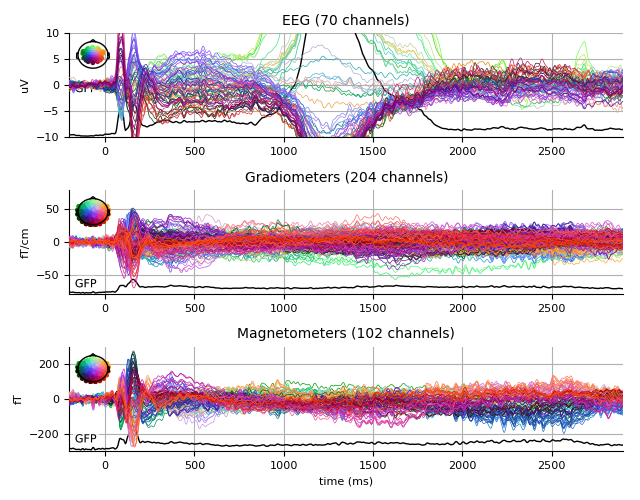
Scrambled
scrambled_evo.plot(spatial_colors=True, gfp=True, ylim=ylim,
window_title='Scrambled %s' % subject)
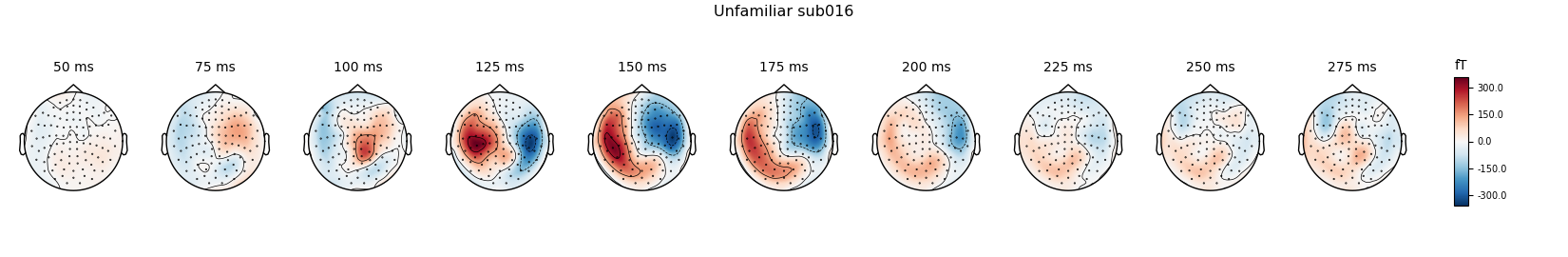
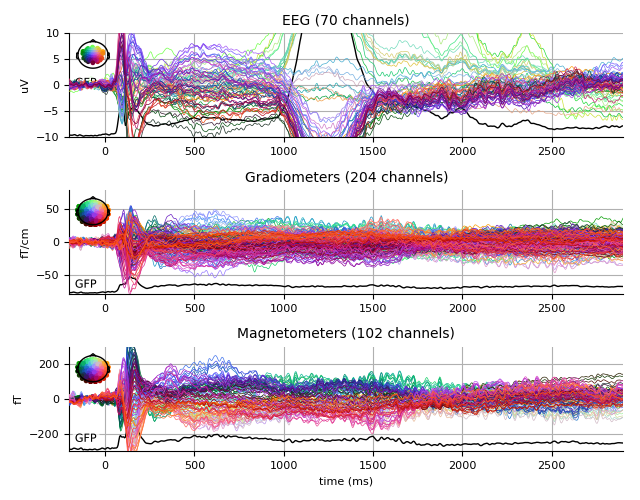
Unfamiliar
unfamiliar_evo.plot(spatial_colors=True, gfp=True, ylim=ylim,
window_title='Unfamiliar %s' % subject)
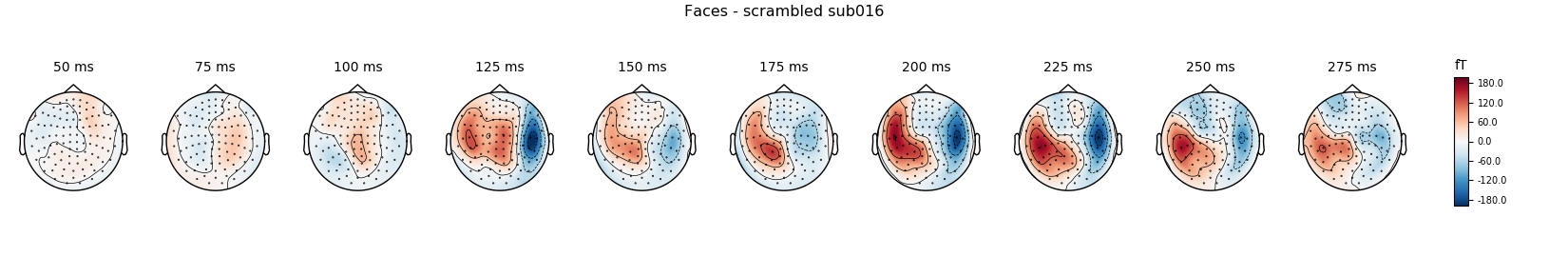
Faces - scrambled
contrast_evo.plot(spatial_colors=True, gfp=True, ylim=ylim,
window_title='Faces - scrambled %s' % subject)
Topomaps
times = np.arange(0.05, 0.3, 0.025)
famous_evo.plot_topomap(times=times, title='Famous %s' % subject,
show=False)
scrambled_evo.plot_topomap(times=times, title='Scrambled %s' % subject,
show=False)
unfamiliar_evo.plot_topomap(times=times, title='Unfamiliar %s' % subject,
show=False)
contrast_evo.plot_topomap(times=times, title='Faces - scrambled %s' % subject,
show=True)
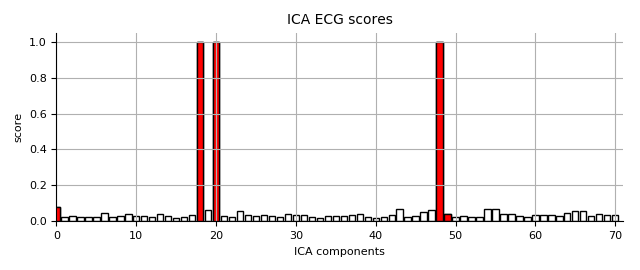
ICA (ECG)
ica_fname = op.join(subject_dir, 'run_concat_highpass-%sHz-ica.fif'
% (l_freq,))
ica = mne.preprocessing.read_ica(ica_fname)
ecg_scores = np.load(
op.join(subject_dir, '%s_highpass-%sHz-ecg-scores.npy'
% (subject, l_freq)))
ica.plot_scores(ecg_scores, show=False, title='ICA ECG scores')
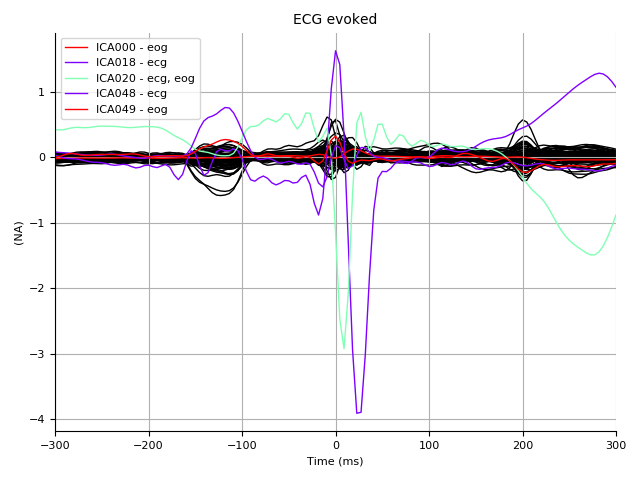
ecg_evoked = mne.read_evokeds(
op.join(subject_dir, '%s_highpass-%sHz-ecg-ave.fif'
% (subject, l_freq)))[0]
ica.plot_sources(ecg_evoked, title='ECG evoked', show=True)
Out:
Reading /tsi/doctorants/data_gramfort/dgw_faces_reproduce/MEG/sub016/run_concat_highpass-NoneHz-ica.fif ...
Now restoring ICA solution ...
Ready.
Reading /tsi/doctorants/data_gramfort/dgw_faces_reproduce/MEG/sub016/sub016_highpass-NoneHz-ecg-ave.fif ...
Read a total of 1 projection items:
Average EEG reference (1 x 70) idle
Found the data of interest:
t = -300.00 ... 300.00 ms (999)
0 CTF compensation matrices available
nave = 3855 - aspect type = 100
Created an SSP operator (subspace dimension = 1)
1 projection items activated
SSP projectors applied...
No baseline correction applied
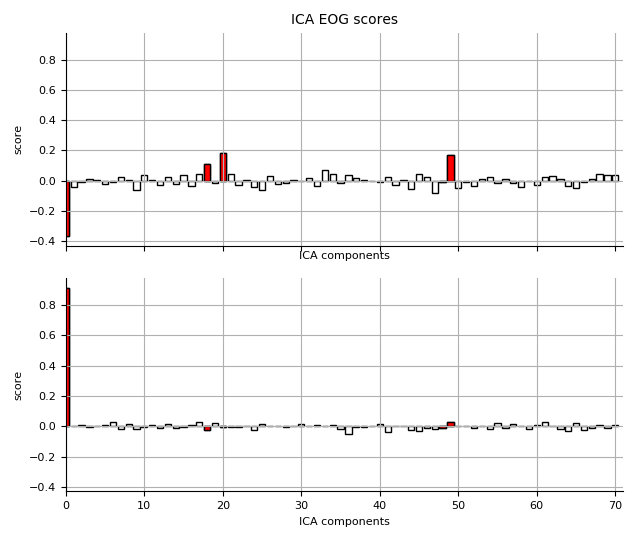
ICA (EOG)
eog_scores = np.load(
op.join(subject_dir, '%s_highpass-%sHz-eog-scores.npy'
% (subject, l_freq)))
ica.plot_scores(eog_scores, show=False, title='ICA EOG scores')
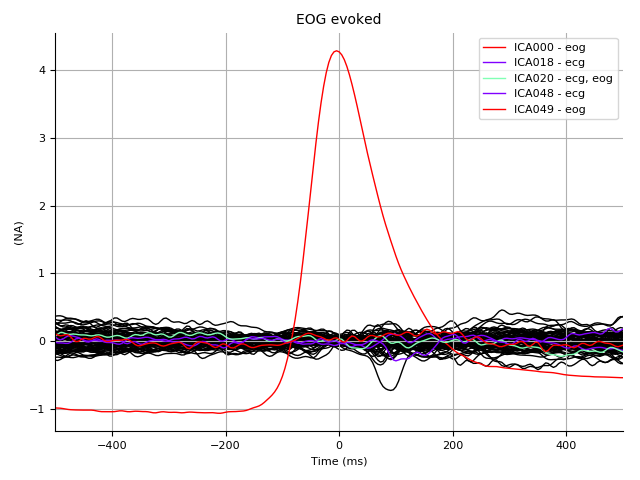
eog_evoked = mne.read_evokeds(
op.join(subject_dir, '%s_highpass-%sHz-eog-ave.fif'
% (subject, l_freq)))[0]
ica.plot_sources(eog_evoked, title='EOG evoked', show=True)
Out:
Reading /tsi/doctorants/data_gramfort/dgw_faces_reproduce/MEG/sub016/sub016_highpass-NoneHz-eog-ave.fif ...
Read a total of 1 projection items:
Average EEG reference (1 x 70) idle
Found the data of interest:
t = -500.00 ... 500.00 ms (998)
0 CTF compensation matrices available
nave = 1212 - aspect type = 100
Created an SSP operator (subspace dimension = 1)
1 projection items activated
SSP projectors applied...
No baseline correction applied
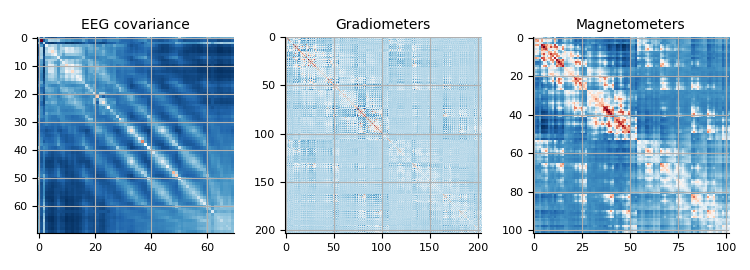
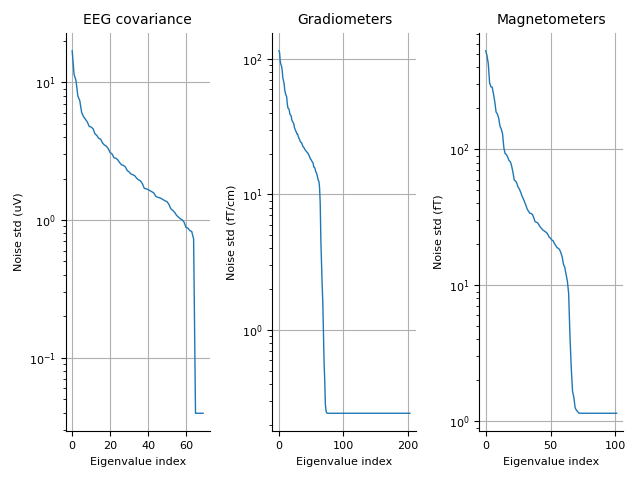
Covariance 07. Evoked data.
cov_fname = op.join(subject_dir,
'%s_highpass-%sHz-cov.fif' % (subject, l_freq))
cov = mne.read_cov(cov_fname)
mne.viz.plot_cov(cov, faces_evo.info)
rank_dict = dict(
meg=raw_filt.copy().load_data().pick_types(eeg=False).estimate_rank())
for kind in ('meg', 'eeg'):
type_dict = dict(meg=False)
type_dict.update({kind: True})
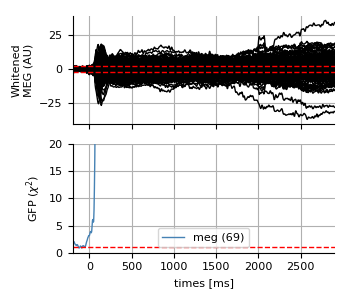
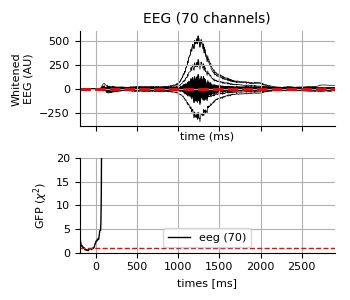
fig = faces_evo.copy().apply_baseline().pick_types(
**type_dict).plot_white(cov, rank=rank_dict if kind == 'meg' else {})
for ax, ylabel in zip(fig.axes, ('Whitened\n%s (AU)' % (kind.upper(),),
'GFP ($\chi^2$)')):
ax.set(ylabel=ylabel)
fig.axes[-1].set(title='', ylim=[0, 20])
fig.axes[-1].legend(loc='lower center')
fig.set_size_inches(3.5, 3, forward=True)
fig.tight_layout()
fig.savefig(op.join('..', 'figures', '%s_highpass-%sHz-plot_white_%s.pdf'
% (subject, l_freq, kind)))
Out:
376 x 376 full covariance (kind = 1) found.
Read a total of 1 projection items:
Average EEG reference (1 x 70) active
Reading 0 ... 545599 = 0.000 ... 495.999 secs...
estimated rank (mag + grad): 69
Applying baseline correction (mode: mean)
SSS has been applied to data. Showing mag and grad whitening jointly.
Setting small MEG eigenvalues to zero.
Not doing PCA for MEG.
Total rank is 69
Applying baseline correction (mode: mean)
estimated rank (eeg): 70
Created an SSP operator (subspace dimension = 1)
Setting small EEG eigenvalues to zero.
Not doing PCA for EEG.
Total rank is 70
TFR 09. Time-frequency decomposition.
fpower = mne.time_frequency.read_tfrs(
op.join(subject_dir, '%s_highpass-%sHz-faces-tfr.h5'
% (subject, l_freq)))[0]
fitc = mne.time_frequency.read_tfrs(
op.join(subject_dir, '%s_highpass-%sHz-itc_faces-tfr.h5'
% (subject, l_freq)))[0]
spower = mne.time_frequency.read_tfrs(
op.join(subject_dir, '%s_highpass-%sHz-scrambled-tfr.h5'
% (subject, l_freq)))[0]
sitc = mne.time_frequency.read_tfrs(
op.join(subject_dir, '%s_highpass-%sHz-itc_scrambled-tfr.h5'
% (subject, l_freq)))[0]
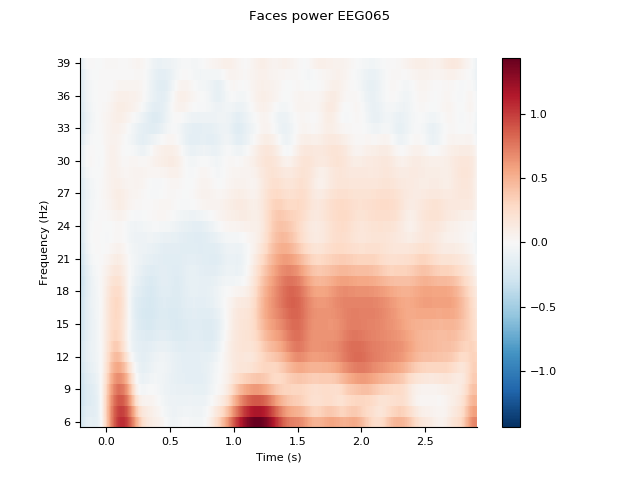
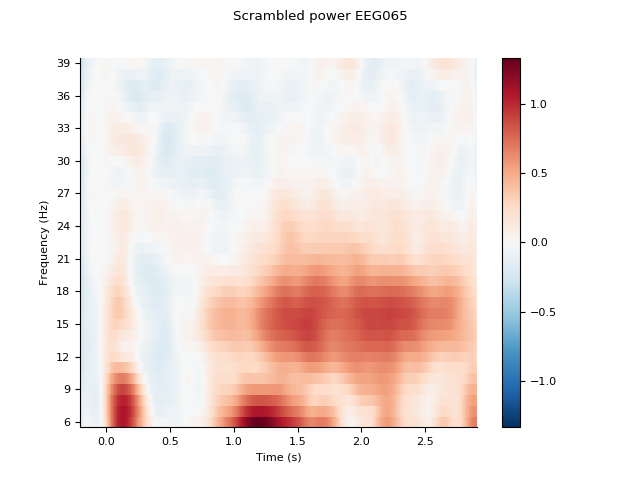
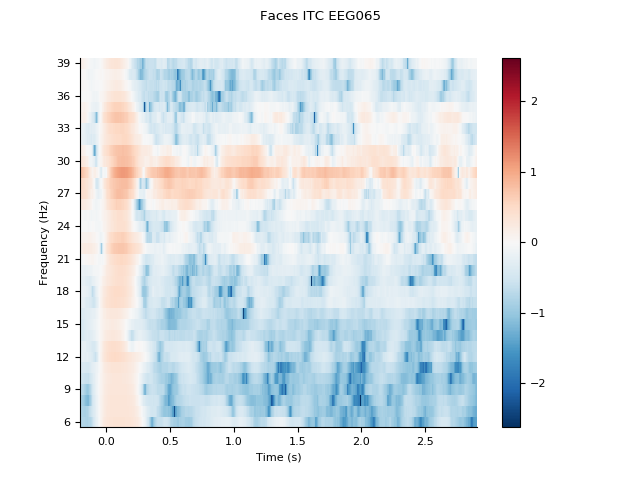
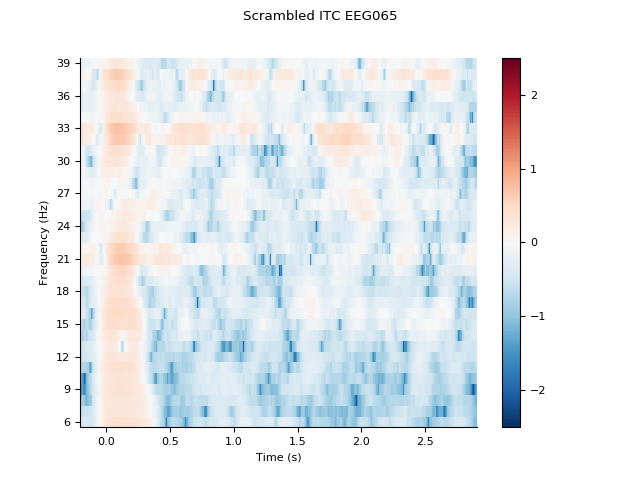
channel = 'EEG065'
idx = [fpower.ch_names.index(channel)]
fpower.plot(idx, title='Faces power %s' % channel, baseline=(-0.1, 0.0),
mode='logratio', show=False)
spower.plot(idx, title='Scrambled power %s' % channel, baseline=(-0.1, 0.0),
mode='logratio', show=False)
fitc.plot(idx, title='Faces ITC %s' % channel, baseline=(-0.1, 0.0),
mode='logratio', show=False)
sitc.plot(idx, title='Scrambled ITC %s' % channel, baseline=(-0.1, 0.0),
mode='logratio')
Out:
Reading /tsi/doctorants/data_gramfort/dgw_faces_reproduce/MEG/sub016/sub016_highpass-NoneHz-faces-tfr.h5 ...
Reading /tsi/doctorants/data_gramfort/dgw_faces_reproduce/MEG/sub016/sub016_highpass-NoneHz-itc_faces-tfr.h5 ...
Reading /tsi/doctorants/data_gramfort/dgw_faces_reproduce/MEG/sub016/sub016_highpass-NoneHz-scrambled-tfr.h5 ...
Reading /tsi/doctorants/data_gramfort/dgw_faces_reproduce/MEG/sub016/sub016_highpass-NoneHz-itc_scrambled-tfr.h5 ...
Applying baseline correction (mode: logratio)
Applying baseline correction (mode: logratio)
Applying baseline correction (mode: logratio)
Applying baseline correction (mode: logratio)
Trans
fname_trans = op.join(study_path, 'ds117', subject, 'MEG',
'%s-trans.fif' % subject)
bem = mne.read_bem_surfaces(op.join(subjects_dir, subject, 'bem',
'%s-5120-bem.fif' % subject))
src = mne.read_source_spaces(
op.join(subjects_dir, subject, 'bem', '%s-oct6-src.fif' % subject))
aln = mne.viz.plot_alignment(
raw.info, fname_trans, subject=subject, subjects_dir=subjects_dir, src=src,
surfaces=['outer_skin', 'inner_skull'], dig=True, coord_frame='meg')
aln.scene.parallel_projection = True
fig, axes = plt.subplots(1, 3, figsize=(6.5, 2.5), facecolor='k')
from mayavi import mlab # noqa: E402
for ai, angle in enumerate((180, 90, 0)):
mlab.view(angle, 90, focalpoint=(0., 0., 0.), distance=0.6)
view = mlab.screenshot()
mask_w = (view == 0).all(axis=-1).all(axis=1)
mask_h = (view == 0).all(axis=-1).all(axis=0)
view = view[~mask_w][:, ~mask_h]
axes[ai].set_axis_off()
axes[ai].imshow(view, interpolation='bicubic')
mlab.close(aln)
fig.subplots_adjust(left=0, right=1, top=1, bottom=0, wspace=0.05, hspace=0)
fig.savefig(op.join('..', 'figures', '%s_alignment.pdf' % subject),
dpi=150, facecolor=fig.get_facecolor(), edgecolor='none')
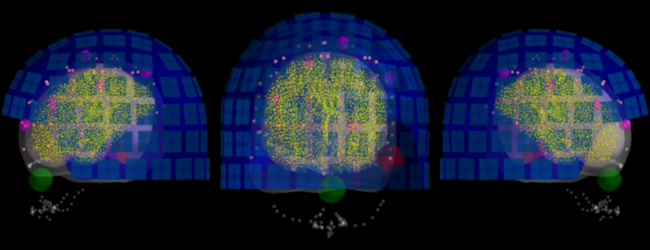
Out:
1 BEM surfaces found
Reading a surface...
[done]
1 BEM surfaces read
Reading a source space...
Computing patch statistics...
Patch information added...
Distance information added...
[done]
Reading a source space...
Computing patch statistics...
Patch information added...
Distance information added...
[done]
2 source spaces read
Using outer_skin for head surface.
Getting helmet for system 306m
Using /tsi/doctorants/data_gramfort/dgw_faces_reproduce/subjects/sub016/bem/flash/inner_skull.surf for head surface.
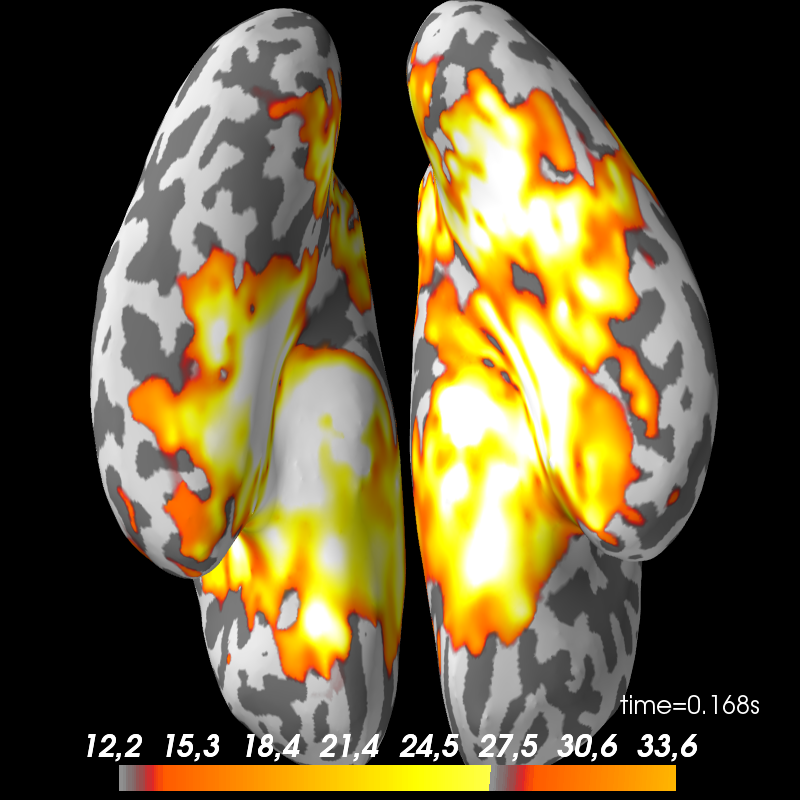
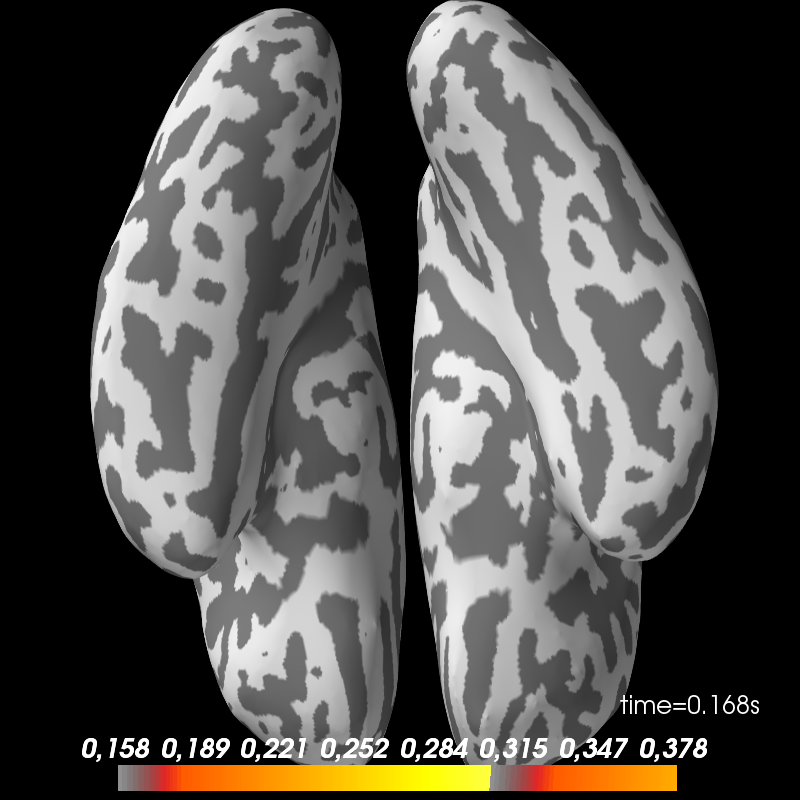
Faces 13. Inverse solution.
def plot_stc(cond, figure=None):
fname = op.join(subject_dir, 'mne_dSPM_inverse_highpass-%sHz-%s'
% (l_freq, cond))
stc = mne.read_source_estimate(fname, subject).magnitude()
brain = stc.plot(subject=subject, subjects_dir=subjects_dir, views=['ven'],
hemi='both', initial_time=0.17, time_unit='s',
figure=figure)
return brain
brain_faces = plot_stc('faces', figure=1)
Faces - scrambled
brain_contrast = plot_stc('contrast', figure=2)
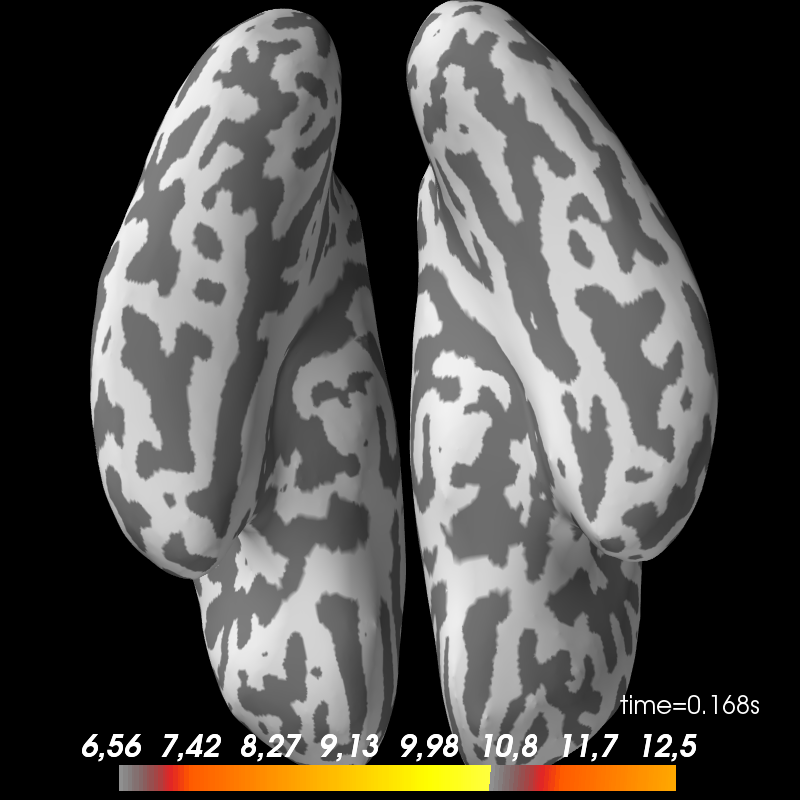
LCMV Faces - scrambled
fname = op.join(subject_dir, 'mne_LCMV_inverse_highpass-%sHz-contrast'
% (l_freq,))
stc = mne.read_source_estimate(fname, subject)
stc.plot(subject=subject, subjects_dir=subjects_dir, views=['ven'],
hemi='both', initial_time=0.17, time_unit='s', figure=3)
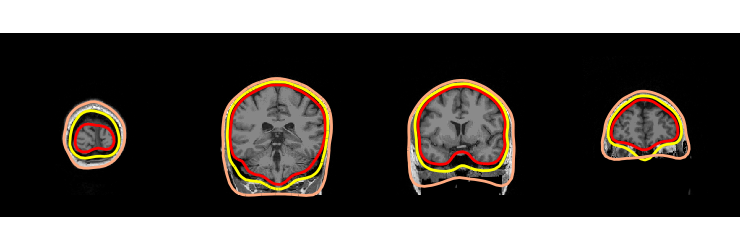
BEM
fig = mne.viz.plot_bem(subject, subjects_dir, slices=[40, 100, 140, 180])
fig.savefig(op.join('..', 'figures', '%s_bem.pdf' % subject))
Out:
Using surface: /tsi/doctorants/data_gramfort/dgw_faces_reproduce/subjects/sub016/bem/inner_skull.surf
Using surface: /tsi/doctorants/data_gramfort/dgw_faces_reproduce/subjects/sub016/bem/outer_skull.surf
Using surface: /tsi/doctorants/data_gramfort/dgw_faces_reproduce/subjects/sub016/bem/outer_skin.surf
Total running time of the script: ( 4 minutes 10.764 seconds)